shortest-path-visualization
If any part of this notebook is used in your research, please cite with the reference found in README.md.
Generating regular lattices and visualizing shortest paths
Creating data for testing and demonstrating routes
Author: James D. Gaboardi jgaboardi@gmail.com
This notebook is a walk-through for:
- Instantiating a simple network through a generated regular lattice
- Generating shortest path geometric objects
- Visualizing shortest paths
%load_ext watermark
%watermark
In addtion to the base spaghetti requirements (and their dependecies), this notebook requires installations of:
- geopandas
$ conda install -c conda-forge geopandas
- matplotlib
$ conda install matplotlib
import spaghetti
import matplotlib
import matplotlib.pyplot as plt
import libpysal
from libpysal.cg import Point
try:
from IPython.display import set_matplotlib_formats
set_matplotlib_formats("retina")
except ImportError:
pass
%matplotlib inline
%watermark -w
%watermark -iv
lattice = spaghetti.regular_lattice(4)
ntw = spaghetti.Network(in_data=lattice)
vertices, arcs = spaghetti.element_as_gdf(ntw, vertices=True, arcs=True)
base = arcs.plot(linewidth=3, alpha=0.25, color="k", zorder=0, figsize=(10, 10))
vertices.plot(ax=base, markersize=20, color="red", zorder=1);
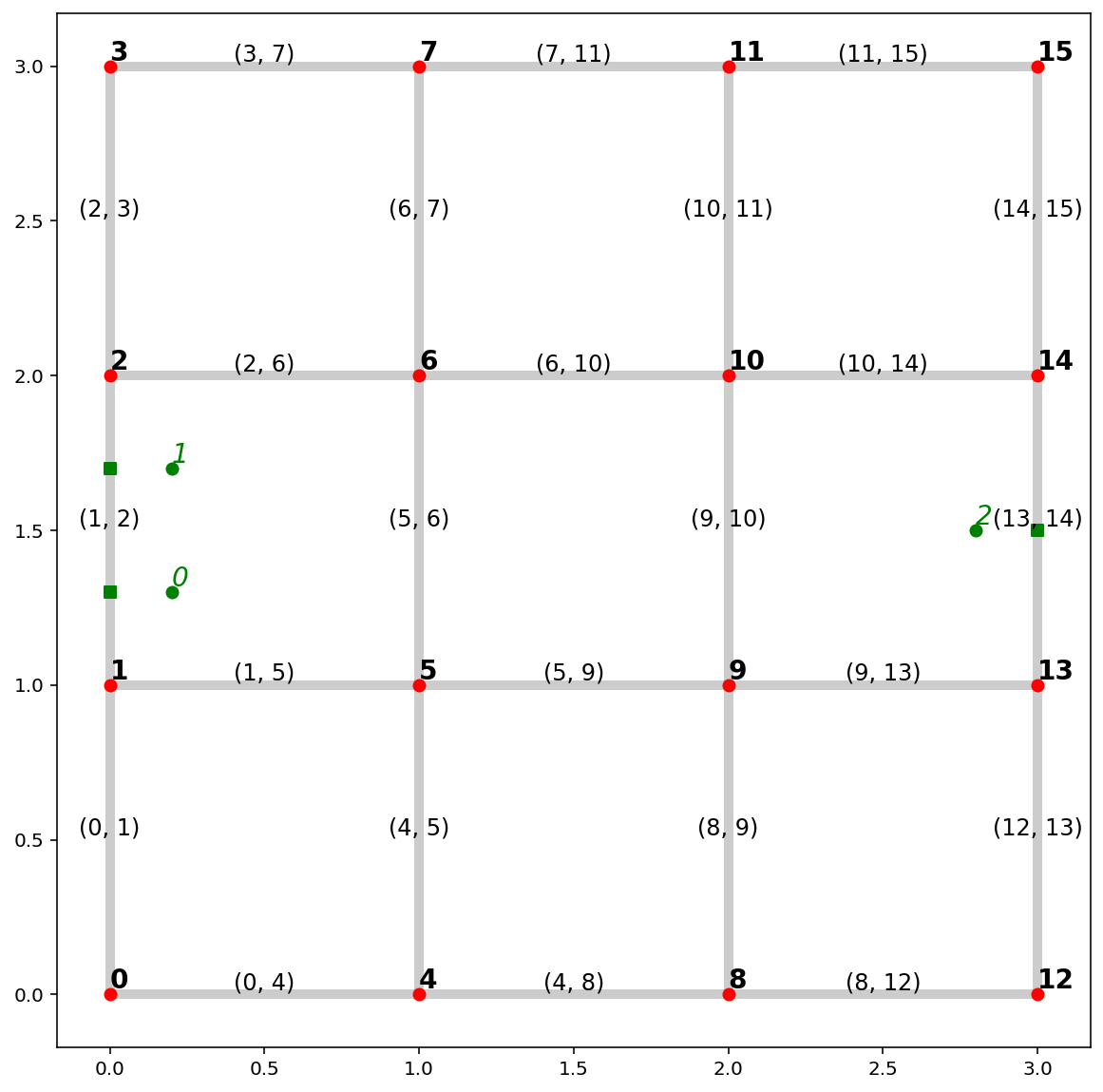
synth_obs = [Point([0.2, 1.3]), Point([0.2, 1.7]), Point([2.8, 1.5])]
ntw.snapobservations(synth_obs, "synth_obs")
# true locations of synthetic observations
pp_obs = spaghetti.element_as_gdf(ntw, pp_name="synth_obs")
# snapped locations of synthetic observations
pp_obs_snapped = spaghetti.element_as_gdf(ntw, pp_name="synth_obs", snapped=True)
base = arcs.plot(alpha=0.2, linewidth=5, color="k", figsize=(10, 10), zorder=0)
vertices.plot(ax=base, color="r", zorder=1)
pp_obs.plot(ax=base, color="g", zorder=2)
pp_obs_snapped.plot(ax=base, color="g", marker="s", zorder=2)
# arc labels
arcs.apply(
lambda x: base.annotate(
s=x.id, xy=x.geometry.centroid.coords[0], size=12, ha="center", va="bottom"
),
axis=1,
)
# vertex labels
vertices.apply(
lambda x: base.annotate(
s=x.id, xy=x.geometry.coords[0], weight="bold", size=14, ha="left", va="bottom"
),
axis=1,
)
# synthetic observation labels
pp_obs.apply(
lambda x: base.annotate(
s=x.id,
xy=x.geometry.coords[0],
style="oblique",
color="g",
size=14,
ha="left",
va="bottom",
),
axis=1,
);
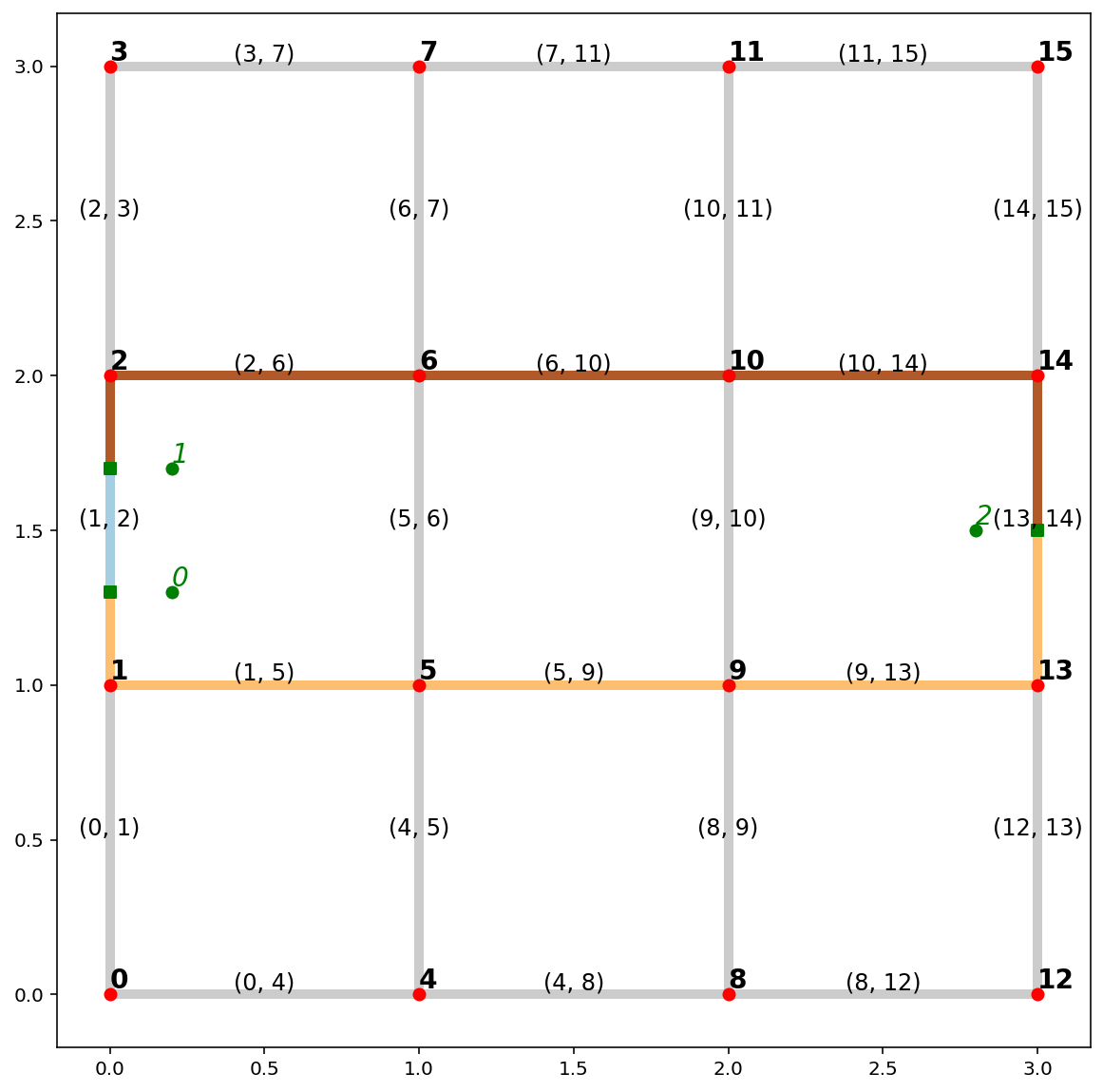
d2d_dist, tree = ntw.allneighbordistances("synth_obs", gen_tree=True)
d2d_dist
tree
paths = ntw.shortest_paths(tree, "synth_obs")
paths
paths_gdf = spaghetti.element_as_gdf(ntw, routes=paths)
paths_gdf
paths_gdf.plot(figsize=(7, 7), column=paths_gdf.index.name, cmap="Paired", linewidth=5);
base = arcs.plot(alpha=0.2, linewidth=5, color="k", figsize=(10, 10), zorder=0)
paths_gdf.plot(ax=base, column="id", cmap="Paired", linewidth=5, zorder=1)
vertices.plot(ax=base, color="r", zorder=2)
pp_obs.plot(ax=base, color="g", zorder=3)
pp_obs_snapped.plot(ax=base, color="g", marker="s", zorder=2)
# arc labels
arcs.apply(
lambda x: base.annotate(
s=x.id, xy=x.geometry.centroid.coords[0], size=12, ha="center", va="bottom"
),
axis=1,
)
# vertex labels
vertices.apply(
lambda x: base.annotate(
s=x.id, xy=x.geometry.coords[0], weight="bold", size=14, ha="left", va="bottom"
),
axis=1,
)
# synthetic observation labels
pp_obs.apply(
lambda x: base.annotate(
s=x.id,
xy=x.geometry.coords[0],
style="oblique",
color="g",
size=14,
ha="left",
va="bottom",
),
axis=1,
);
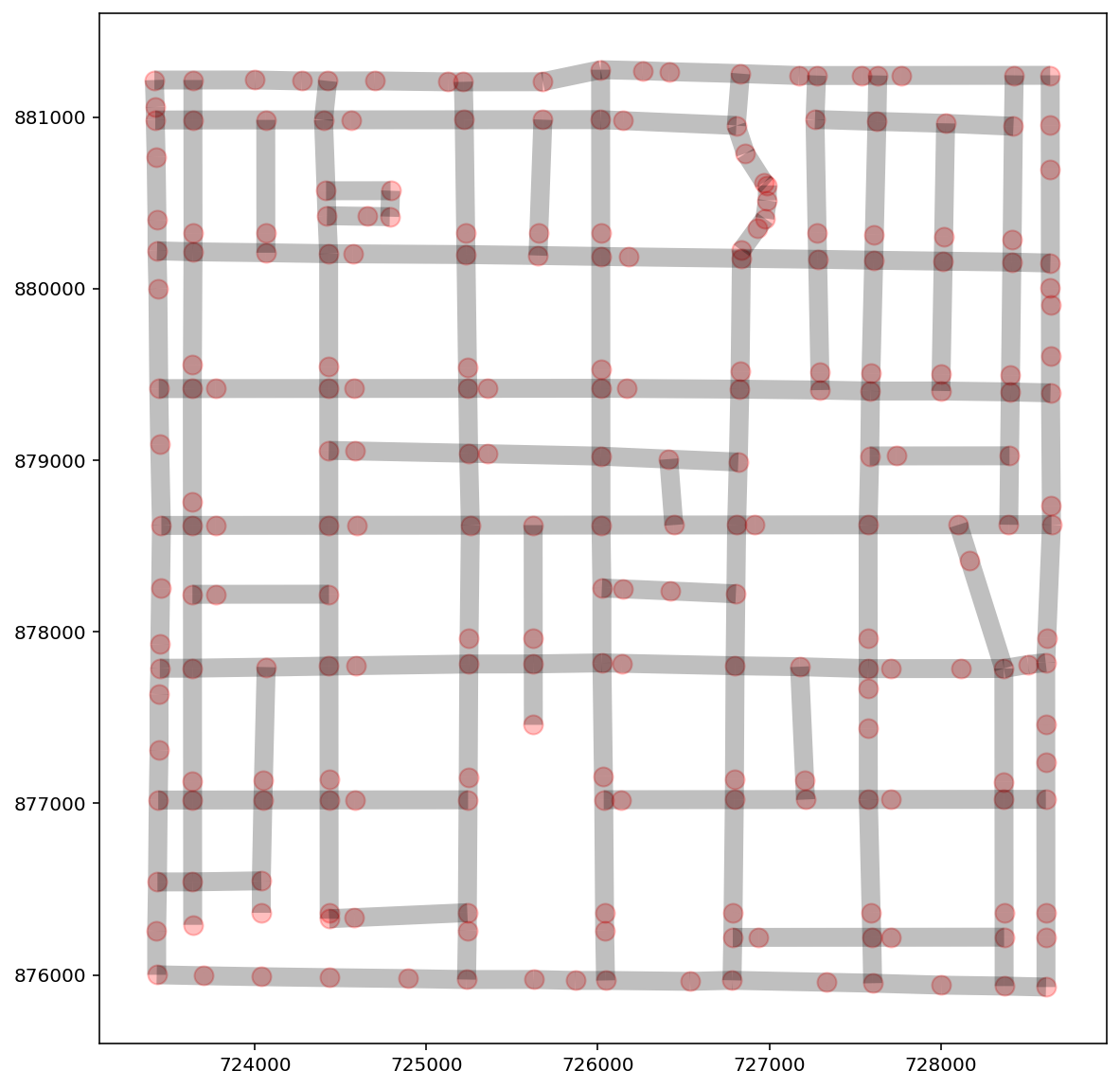
ntw = spaghetti.Network(in_data=libpysal.examples.get_path("streets.shp"))
vertices, arcs = spaghetti.element_as_gdf(ntw, vertices=True, arcs=True)
base = arcs.plot(linewidth=10, alpha=0.25, color="k", figsize=(10, 10))
vertices.plot(ax=base, markersize=100, alpha=0.25, color="red");
ntw.snapobservations(libpysal.examples.get_path("schools.shp"), "schools")
pp_obs = spaghetti.element_as_gdf(ntw, pp_name="schools")
pp_obs_snapped = spaghetti.element_as_gdf(ntw, pp_name="schools", snapped=True)
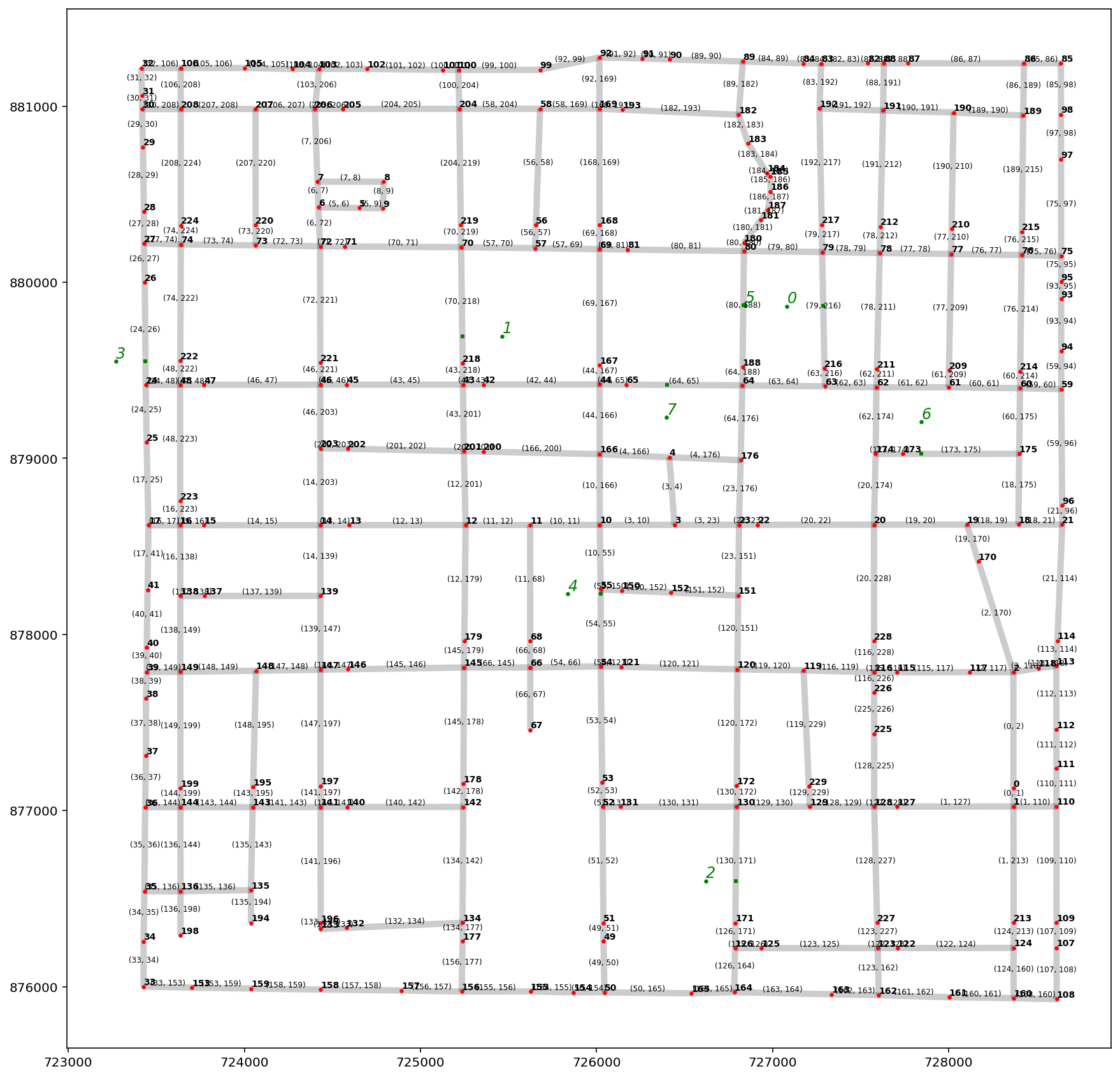
base = arcs.plot(alpha=0.2, linewidth=5, color="k", figsize=(15, 15), zorder=0)
vertices.plot(ax=base, markersize=5, color="r", zorder=1)
pp_obs.plot(ax=base, markersize=5, color="g", zorder=2)
pp_obs_snapped.plot(ax=base, markersize=5, marker="s", color="g", zorder=2)
# arc labels
arcs.apply(
lambda x: base.annotate(
s=x.id, xy=x.geometry.centroid.coords[0], size=6, ha="center", va="bottom"
),
axis=1,
)
# vertex labels
vertices.apply(
lambda x: base.annotate(
s=x.id, xy=x.geometry.coords[0], weight="bold", size=7, ha="left", va="bottom"
),
axis=1,
)
# school labels
pp_obs.apply(
lambda x: base.annotate(
s=x.id,
xy=x.geometry.coords[0],
style="oblique",
color="g",
size=12,
ha="left",
va="bottom",
),
axis=1,
);
d2d_dist, tree = ntw.allneighbordistances("schools", gen_tree=True)
paths = ntw.shortest_paths(tree, "schools")
paths_gdf = spaghetti.element_as_gdf(ntw, routes=paths)
paths_gdf.head()
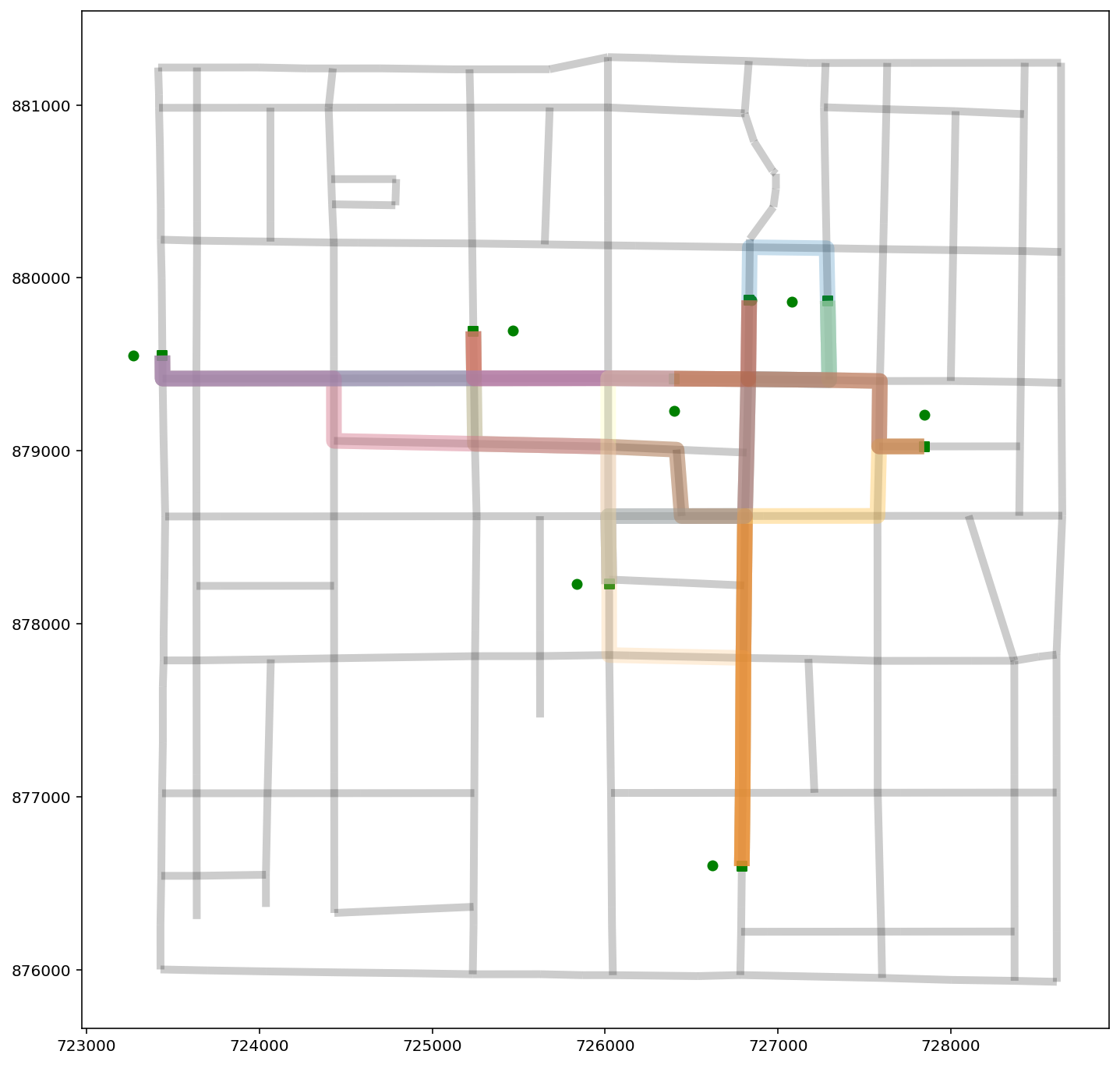
paths_gdf.plot(figsize=(7, 7), column=paths_gdf.index.name, cmap="Paired", linewidth=5);
base = arcs.plot(alpha=0.2, linewidth=5, color="k", figsize=(12, 12), zorder=0)
pp_obs.plot(ax=base, color="g", zorder=2)
pp_obs_snapped.plot(ax=base, color="g", marker="s", zorder=2)
paths_gdf.plot(ax=base, column="id", cmap="Paired", linewidth=10, alpha=0.25);