network-analysis
If any part of this notebook is used in your research, please cite with the reference found in README.md.
Spatial network analysis
Demonstrating network representation and cluster detection
Author: James D. Gaboardi jgaboardi@gmail.com
This notebook is an advanced walk-through for:
- Exploring the attributes of network objects and point patterns
- Understanding the difference between a network and its graph-theoretic representation
- Performing spatial network analysis
%load_ext watermark
%watermark
In addtion to the base spaghetti requirements (and their dependecies), this notebook requires installations of:
- matplotlib
$ conda install matplotlib
import spaghetti
import esda
import libpysal
import matplotlib
import matplotlib.pyplot as plt
import numpy
import warnings
try:
from IPython.display import set_matplotlib_formats
set_matplotlib_formats("retina")
except ImportError:
pass
%matplotlib inline
%watermark -w
%watermark -iv
ntw = spaghetti.Network(in_data=libpysal.examples.get_path("streets.shp"))
# Crimes
ntw.snapobservations(
libpysal.examples.get_path("crimes.shp"), "crimes", attribute=True
)
# Schools
ntw.snapobservations(
libpysal.examples.get_path("schools.shp"), "schools", attribute=False
)
ntw.pointpatterns
Attributes for every point pattern
dist_snappeddict keyed by point id with the value as snapped distance from observation to network arc
ntw.pointpatterns["crimes"].dist_snapped[0]
dist_to_vertexdict keyed by pointid with the value being a dict in the form{node: distance to vertex, node: distance to vertex}
ntw.pointpatterns["crimes"].dist_to_vertex[0]
npointspoint observations in set
ntw.pointpatterns["crimes"].npoints
obs_to_arcdict keyed by arc with the value being a dict in the form{pointID:(x-coord, y-coord), pointID:(x-coord, y-coord), ... }
ntw.pointpatterns["crimes"].obs_to_arc[(161, 162)]
obs_to_vertexlist of incident network vertices to snapped observation points
ntw.pointpatterns["crimes"].obs_to_vertex[0]
pointsgeojson like representation of the point pattern. Includes properties if read with attributes=True
ntw.pointpatterns["crimes"].points[0]
snapped_coordinatesdict keyed by pointid with the value being (x-coord, y-coord)
ntw.pointpatterns["crimes"].snapped_coordinates[0]
Counts per link (arc or edge) are important, but should not be precomputed since we have different representations of the network (spatial and graph currently). (Relatively) Uniform segmentation still needs to be done.
counts = ntw.count_per_link(ntw.pointpatterns["crimes"].obs_to_arc, graph=False)
orig_counts_per_link = sum(list(counts.values())) / float(len(counts.keys()))
orig_counts_per_link
n200 = ntw.split_arcs(200.0)
counts = n200.count_per_link(n200.pointpatterns["crimes"].obs_to_arc, graph=False)
segm_counts_per_link = sum(counts.values()) / float(len(counts.keys()))
segm_counts_per_link
# 'full' unsegmented network
vertices_df, arcs_df = spaghetti.element_as_gdf(
ntw, vertices=ntw.vertex_coords, arcs=ntw.arcs
)
# network segmented at 200-meter increments
vertices200_df, arcs200_df = spaghetti.element_as_gdf(
n200, vertices=n200.vertex_coords, arcs=n200.arcs
)
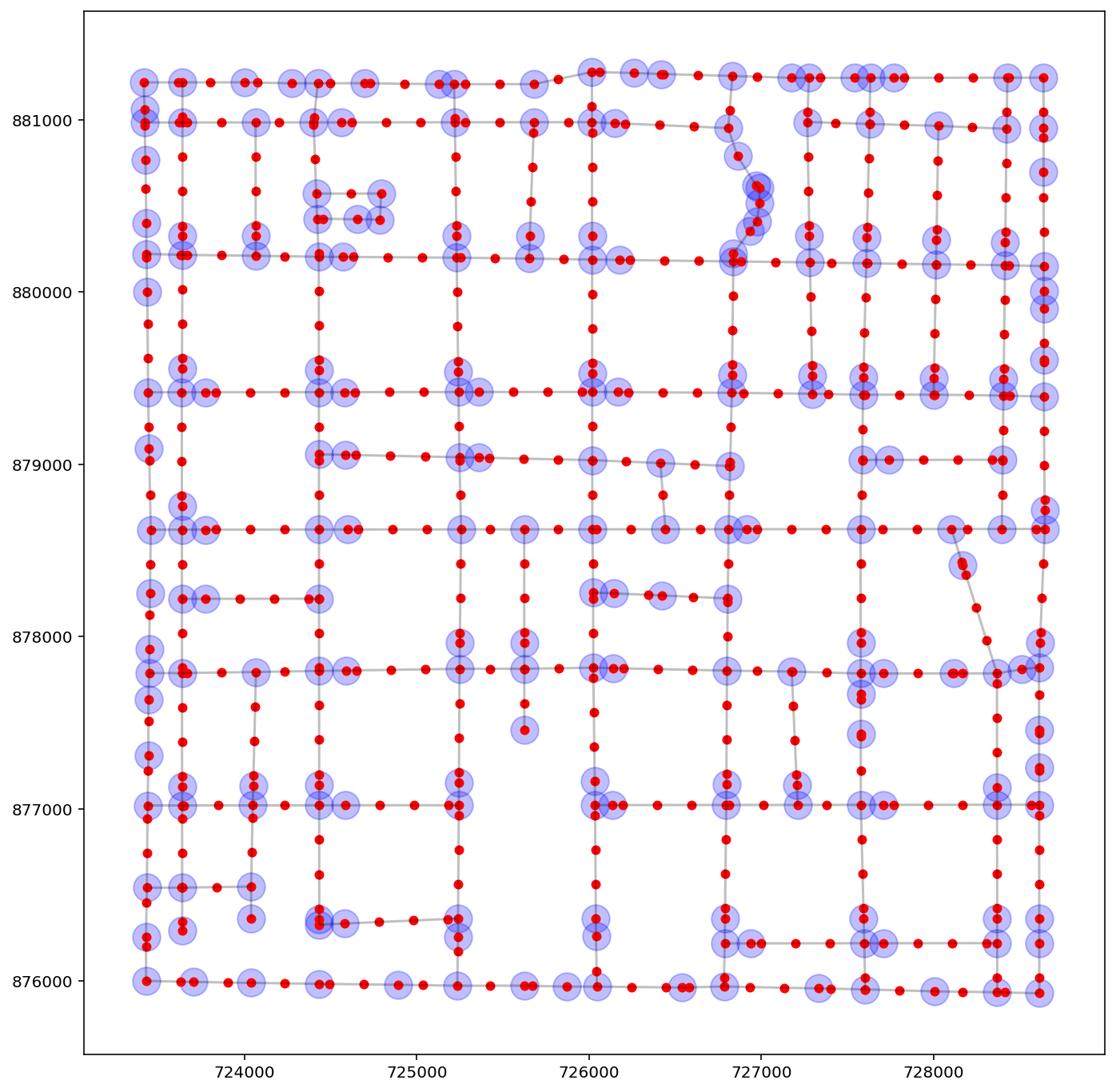
base = arcs_df.plot(color="k", alpha=0.25, figsize=(12, 12))
vertices_df.plot(ax=base, color="b", markersize=300, alpha=0.25)
vertices200_df.plot(ax=base, color="r", markersize=25, alpha=1.0);
# Compute the counts
counts = ntw.count_per_link(ntw.pointpatterns["crimes"].obs_to_arc, graph=False)
# Binary Adjacency
w = ntw.contiguityweights(graph=False)
# Build the y vector
arcs = w.neighbors.keys()
y = numpy.zeros(len(arcs))
for i, a in enumerate(arcs):
if a in counts.keys():
y[i] = counts[a]
# Moran's I
res = esda.moran.Moran(y, w, permutations=99)
print(dir(res))
# Compute the counts
counts = ntw.count_per_link(ntw.pointpatterns["crimes"].obs_to_arc, graph=True)
# Binary Adjacency
w = ntw.contiguityweights(graph=True)
# Build the y vector
edges = w.neighbors.keys()
y = numpy.zeros(len(edges))
for i, e in enumerate(edges):
if e in counts.keys():
y[i] = counts[e]
# Moran's I
res = esda.moran.Moran(y, w, permutations=99)
print(dir(res))
# Compute the counts
counts = n200.count_per_link(n200.pointpatterns["crimes"].obs_to_arc, graph=False)
# Binary Adjacency
w = n200.contiguityweights(graph=False)
# Build the y vector and convert from raw counts to intensities
arcs = w.neighbors.keys()
y = numpy.zeros(len(n200.arcs))
for i, a in enumerate(arcs):
if a in counts.keys():
length = n200.arc_lengths[a]
y[i] = float(counts[a]) / float(length)
# Moran's I
res = esda.moran.Moran(y, w, permutations=99)
print(dir(res))
npts = ntw.pointpatterns["crimes"].npoints
sim = ntw.simulate_observations(npts)
print(dir(sim))
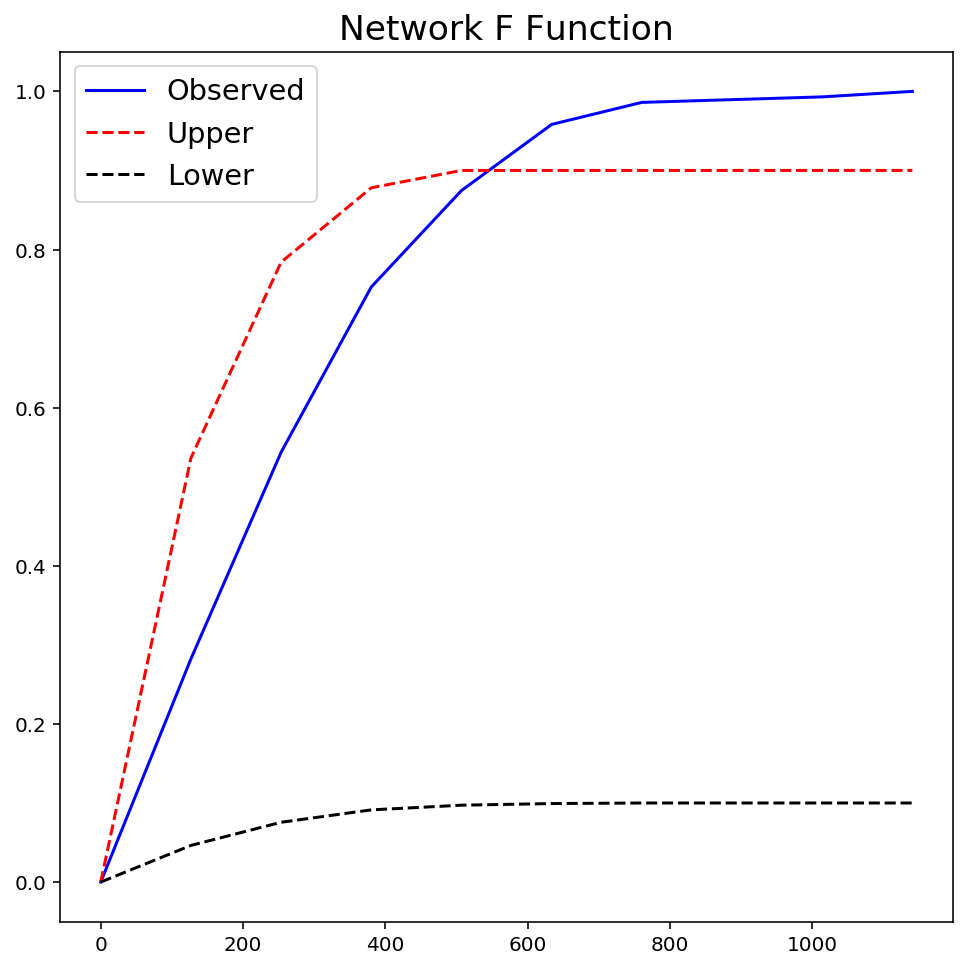
fres = ntw.NetworkF(ntw.pointpatterns["crimes"], permutations=99)
plt.figure(figsize=(8, 8))
plt.plot(fres.xaxis, fres.observed, "b-", linewidth=1.5, label="Observed")
plt.plot(fres.xaxis, fres.upperenvelope, "r--", label="Upper")
plt.plot(fres.xaxis, fres.lowerenvelope, "k--", label="Lower")
plt.legend(loc="best", fontsize="x-large")
plt.title("Network F Function", fontsize="xx-large")
plt.show()
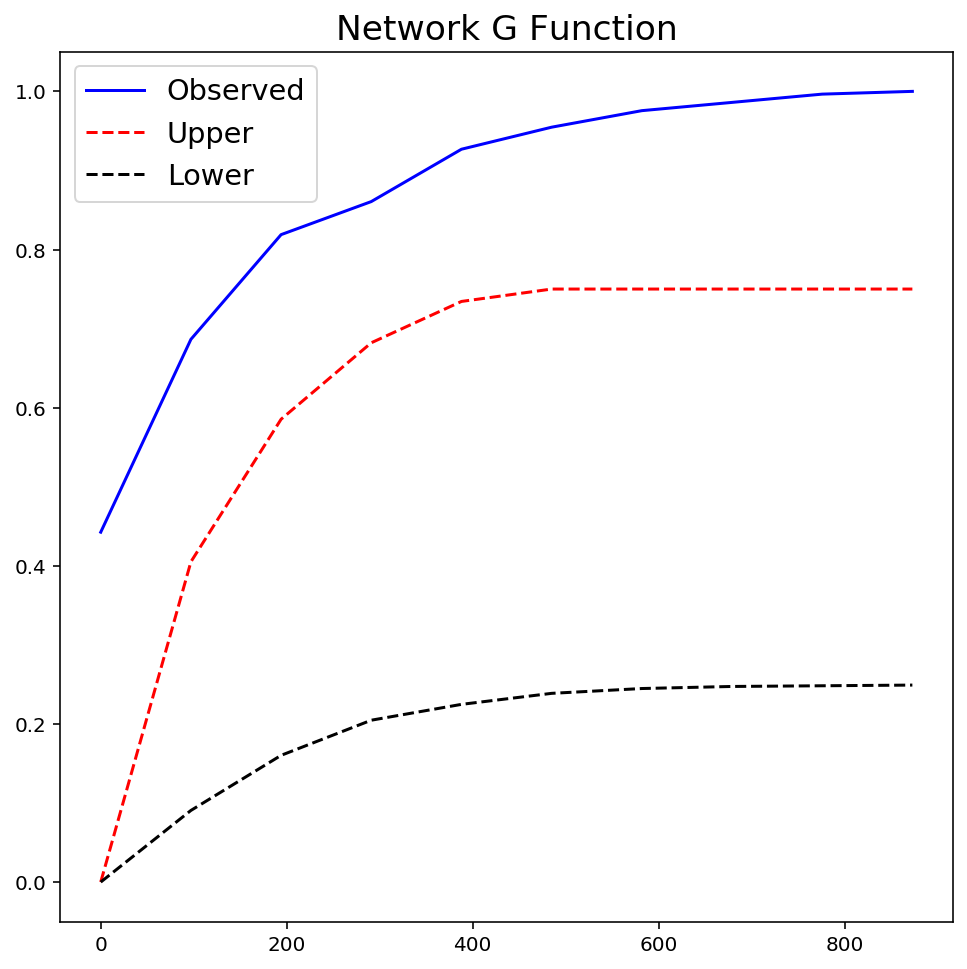
gres = ntw.NetworkG(ntw.pointpatterns["crimes"], permutations=99)
plt.figure(figsize=(8, 8))
plt.plot(gres.xaxis, gres.observed, "b-", linewidth=1.5, label="Observed")
plt.plot(gres.xaxis, gres.upperenvelope, "r--", label="Upper")
plt.plot(gres.xaxis, gres.lowerenvelope, "k--", label="Lower")
plt.legend(loc="best", fontsize="x-large")
plt.title("Network G Function", fontsize="xx-large")
plt.show()
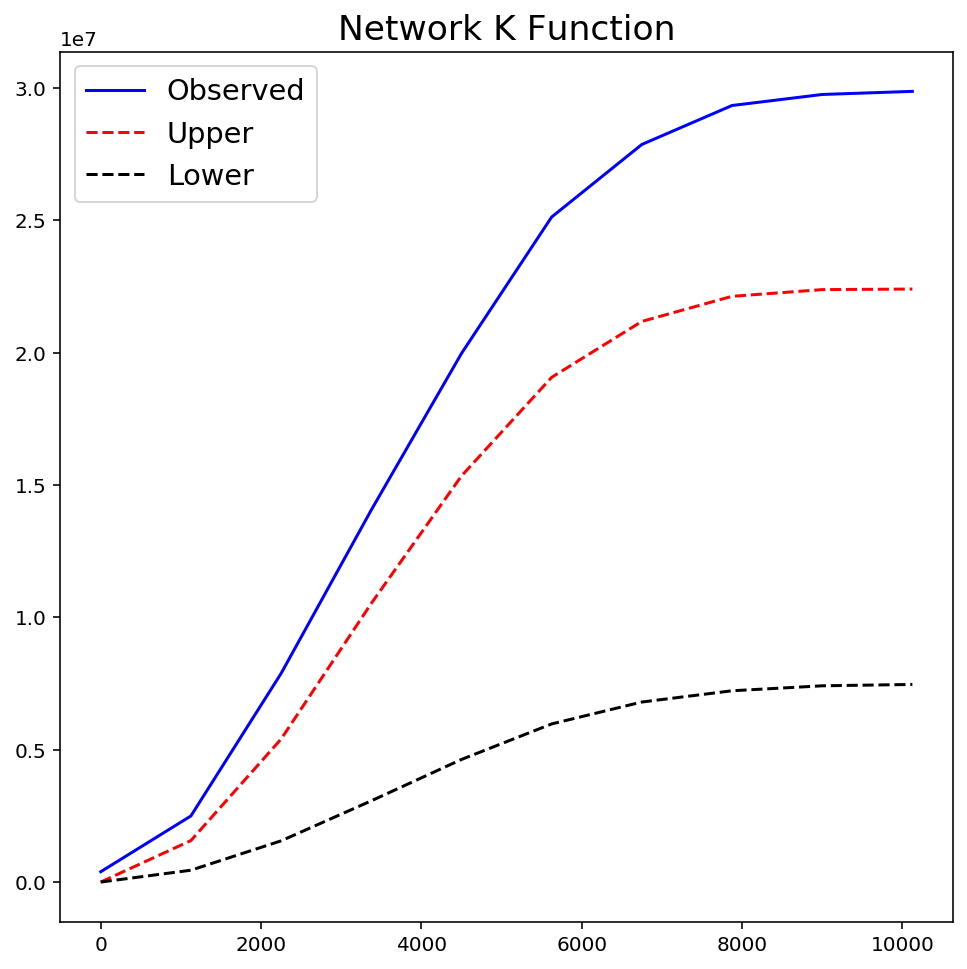
warnings.filterwarnings("ignore", message="invalid value encountered")
kres = ntw.NetworkK(ntw.pointpatterns["crimes"], permutations=99)
plt.figure(figsize=(8, 8))
plt.plot(kres.xaxis, kres.observed, "b-", linewidth=1.5, label="Observed")
plt.plot(kres.xaxis, kres.upperenvelope, "r--", label="Upper")
plt.plot(kres.xaxis, kres.lowerenvelope, "k--", label="Lower")
plt.legend(loc="best", fontsize="x-large")
plt.title("Network K Function", fontsize="xx-large")
plt.show()