connected-components
If any part of this notebook is used in your research, please cite with the reference found in README.md.
Connected components in a spatial network
Identifying and visualizing the parts of a network
Author: James D. Gaboardi jgaboardi@gmail.com
This notebook is a walk-through for:
- Instantiating a simple network with
libpysal.cg.Chainobjects - Working with the network components
- Visualizing the components and (non)articulation vertices
%load_ext watermark
%watermark
In addtion to the base spaghetti requirements (and their dependecies), this notebook requires installations of:
- geopandas
$ conda install -c conda-forge geopandas
- matplotlib
$ conda install matplotlib
import spaghetti
import geopandas
import libpysal
from libpysal.cg import Point, Chain
import matplotlib
try:
from IPython.display import set_matplotlib_formats
set_matplotlib_formats("retina")
except ImportError:
pass
%matplotlib inline
%watermark -w
%watermark -iv
plus1 = [
Chain([Point([1, 2]), Point([0, 2])]),
Chain([Point([1, 2]), Point([1, 1])]),
Chain([Point([1, 2]), Point([1, 3])]),
]
plus2 = [
Chain([Point([2, 1]), Point([2, 0])]),
Chain([Point([2, 1]), Point([3, 1])]),
Chain([Point([2, 1]), Point([2, 2])]),
]
lines = plus1 + plus2
ntw = spaghetti.Network(in_data=lines)
ntw.network_n_components
ntw.network_component2arc
# network vertices and arcs
vertices_df, arcs_df = spaghetti.element_as_gdf(ntw, vertices=True, arcs=True)
arcs_df
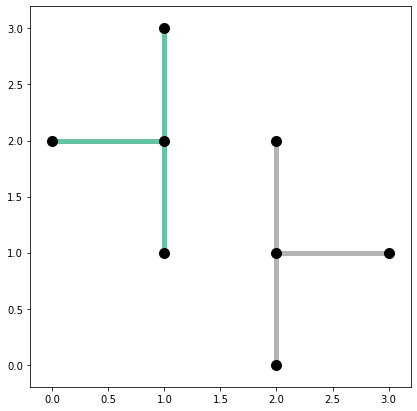
base = arcs_df.plot(column="comp_label", cmap="Set2", linewidth=5, figsize=(7, 7))
vertices_df.plot(ax=base, color="k", markersize=100, zorder=2);
new_lines = [
Chain([Point([1, 1]), Point([2, 2])]),
Chain([Point([0.5, 1]), Point([0.5, 0.5])]),
Chain([Point([0.5, 0.5]), Point([1, 0.5])]),
Chain([Point([2, 2.5]), Point([2.5, 2.5])]),
Chain([Point([2.5, 2.5]), Point([2.5, 2])]),
]
lines += new_lines
ntw = spaghetti.Network(in_data=lines)
ntw.network_n_components
ntw.network_component2arc
# network vertices and arcs
vertices_df, arcs_df = spaghetti.element_as_gdf(ntw, vertices=True, arcs=True)
arcs_df
ntw.non_articulation_points
napts = ntw.non_articulation_points
articulation_vertices = vertices_df[~vertices_df["id"].isin(napts)]
non_articulation_vertices = vertices_df[vertices_df["id"].isin(napts)]
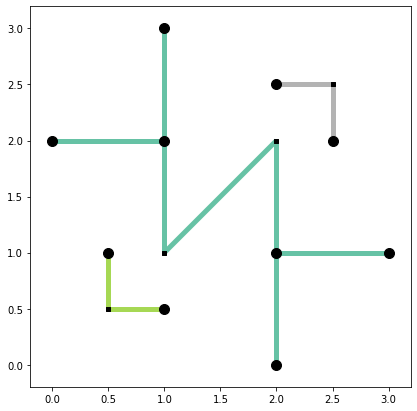
base = arcs_df.plot(column="comp_label", cmap="Set2", linewidth=5, figsize=(7, 7))
articulation_vertices.plot(ax=base, color="k", markersize=100, zorder=2)
non_articulation_vertices.plot(ax=base, marker="s", color="k", markersize=20, zorder=2);
new_lines = [
Chain([Point([3, 1]), Point([3.25, 1.25])]),
Chain([Point([3.25, 1.25]), Point([3.5, 1.25])]),
Chain([Point([3.5, 1.25]), Point([3.75, 1])]),
Chain([Point([3.75, 1]), Point([3.5, .75])]),
Chain([Point([3.5, .75]), Point([3.25, .75])]),
Chain([Point([3.25, .75]), Point([3, 1])]),
]
lines += new_lines
ntw = spaghetti.Network(in_data=lines)
ntw.network_n_components
ntw.network_component2arc
# network vertices and arcs
vertices_df, arcs_df = spaghetti.element_as_gdf(ntw, vertices=True, arcs=True)
arcs_df
ntw.non_articulation_points
napts = ntw.non_articulation_points
articulation_vertices = vertices_df[~vertices_df["id"].isin(napts)]
non_articulation_vertices = vertices_df[vertices_df["id"].isin(napts)]
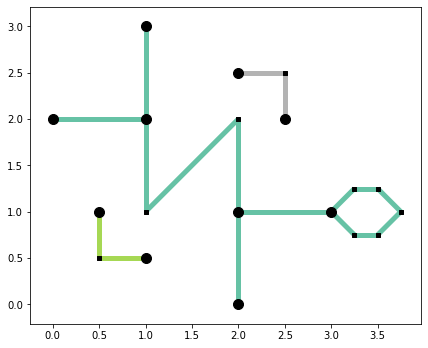
base = arcs_df.plot(column="comp_label", cmap="Set2", linewidth=5, figsize=(7, 7))
articulation_vertices.plot(ax=base, color="k", markersize=100, zorder=2)
non_articulation_vertices.plot(ax=base, marker="s", color="k", markersize=20, zorder=2);