This page was generated from notebooks/p-median_variations.ipynb.
Interactive online version:
Comparing P-Median Variations¶
This tutorial compares 4 versions of the \(p\)-median problem.
Authors
Contents
Model data set up & inputs
The classic \(p\)-median problem
The capacited \(p\)-median problem
The \(k\)-nearest \(p\)-median problem
The capacited \(k\)-nearest \(p\)-median problem
Comparing variant results
References
Notes
All models are solved with euclidean distance as impedence
Detailed explanations of the classic \(p\)-median problem and the capacited variant can be found in the p-median tutorial. These models were included in
spoptwith GSoC 2021 by Germano Barcelos and AGILE 2023 by Rongbo Xu, respectively.The \(k\)-nearest \(p\)-median problem implementations were part of GSoC 2023 and contributed by Rongbo Xu. The complete, detailed write-up can be found here.
[1]:
%config InlineBackend.figure_format = "retina"
%load_ext watermark
%watermark
Last updated: 2025-04-07T15:16:33.243843-04:00
Python implementation: CPython
Python version : 3.12.9
IPython version : 9.0.2
Compiler : Clang 18.1.8
OS : Darwin
Release : 24.4.0
Machine : arm64
Processor : arm
CPU cores : 8
Architecture: 64bit
[2]:
import geopandas
import matplotlib.colors as mcolors
import matplotlib.lines as mlines
import matplotlib.pyplot as plt
import numpy
import pandas
import pulp
import shapely
from matplotlib.patches import Patch
import spopt
from spopt.locate import KNearestPMedian, PMedian, simulated_geo_points
%watermark -w
%watermark -iv
Watermark: 2.5.0
pandas : 2.2.3
geopandas : 1.0.1
numpy : 2.2.4
matplotlib: 3.10.1
spopt : 0.6.2.dev3+g13ca45e
shapely : 2.1.0
pulp : 2.8.0
1. Model data set up & inputs¶
Constants¶
N_CLI: The quantity of client locations to simulateN_FAC: The quantity of facility locations to simulateP_FAC: Candidate facilities in an optimal solutionSEED_CLI/SEED_FAC: Random state seeds for reproducibilitysolver: The solver engine to utilize for optimization
[3]:
N_CLI = 100
N_FAC = 100
P_FAC = 8
SEED_CLI = 7
SEED_FAC = 5
solver = pulp.COIN_CMD(msg=False, warmStart=True)
Simulate client and candidate facility locations¶
The simulated_geo_points utility function available in spopt simulates points in a study area.
[4]:
study_area = shapely.Polygon(((0, 0), (10, 0), (10, 10), (0, 10)))
clients = simulated_geo_points(study_area, needed=N_CLI, seed=SEED_CLI)
facilities = simulated_geo_points(study_area, needed=N_FAC, seed=SEED_FAC)
for df in [clients, facilities]:
df.set_crs("EPSG:4326", inplace=True)
[5]:
fig, ax = plt.subplots(figsize=(4, 4))
_label = f"client sites ($n$={N_CLI})"
clients.plot(ax=ax, label=_label, alpha=0.3)
_label = f"facility candidate sites ($n$={N_FAC})"
facilities.plot(ax=ax, zorder=2, label=_label, alpha=0.3)
plt.legend(loc="upper left", bbox_to_anchor=(1.05, 1));
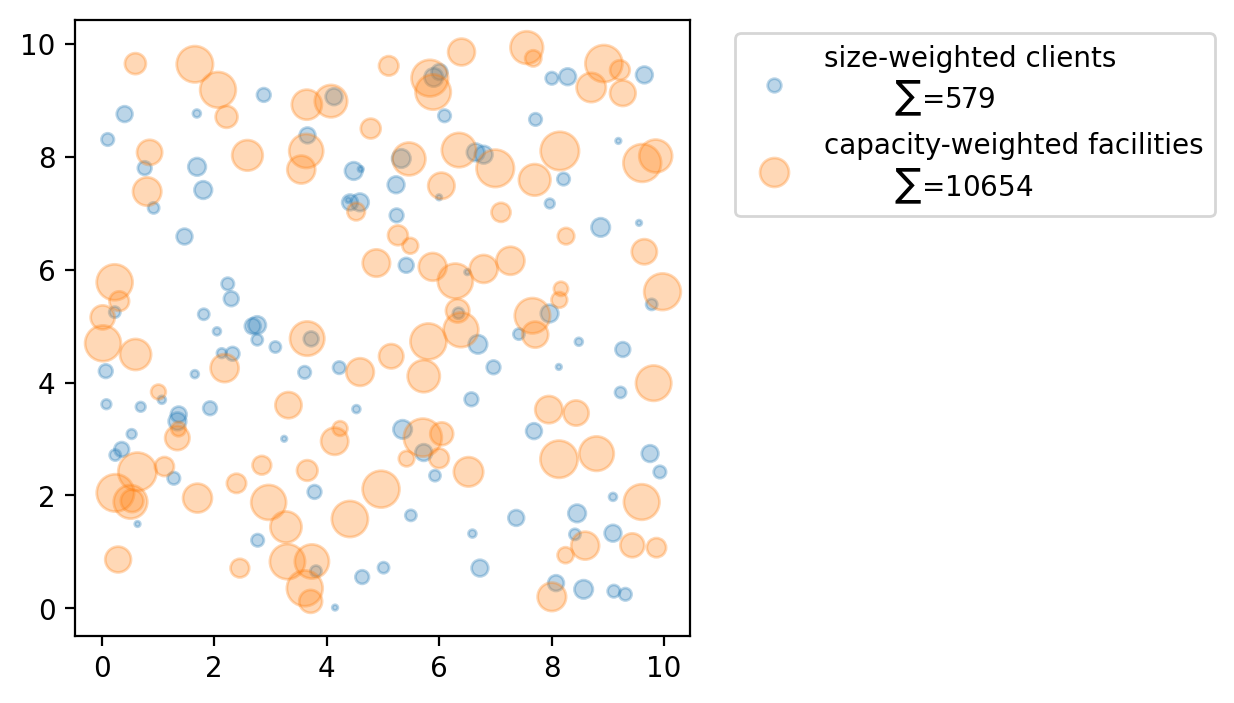
For each client location all \(p\)-median model varinates suppose there is a weight. Here we assign random integer values using numpy to simulate these weights ranging from a minimum of 1 and a maximum of 12.
[6]:
numpy.random.seed(0)
ai = numpy.random.randint(1, 12, N_CLI)
clients["weights"] = ai
sum_clients = clients["weights"].sum()
print(f"The total of client weights: {sum_clients}")
clients
The total of client weights: 579
[6]:
| geometry | weights | |
|---|---|---|
| 0 | POINT (0.76308 7.79919) | 6 |
| 1 | POINT (4.38409 7.23465) | 1 |
| 2 | POINT (9.7799 5.38496) | 4 |
| 3 | POINT (5.0112 0.72051) | 4 |
| 4 | POINT (2.68439 4.99883) | 8 |
| ... | ... | ... |
| 95 | POINT (1.06877 3.69486) | 2 |
| 96 | POINT (2.32671 4.51079) | 6 |
| 97 | POINT (2.76317 5.01807) | 10 |
| 98 | POINT (9.22603 3.82511) | 4 |
| 99 | POINT (6.50128 5.95621) | 1 |
100 rows × 2 columns
For each service facility the :math:`p`-median capacitated variants suppose there is a service level available at each site (the capacity). Again, we assign random integer values using numpy to simulate these capacities ranging from a minimum of 50 and a maximum of 200.
[7]:
min_cap = 50
max_cap = 200
numpy.random.seed(1)
cj = numpy.random.randint(min_cap, max_cap, N_FAC)
facilities["capacity"] = cj
sum_capacity = facilities["capacity"].sum()
print(f"The total of service capacity: {sum_capacity}")
facilities
The total of service capacity: 12126
[7]:
| geometry | capacity | |
|---|---|---|
| 0 | POINT (2.21993 8.70732) | 87 |
| 1 | POINT (2.06719 9.18611) | 190 |
| 2 | POINT (4.88411 6.11744) | 122 |
| 3 | POINT (7.65908 5.18418) | 187 |
| 4 | POINT (2.96801 1.87721) | 183 |
| ... | ... | ... |
| 95 | POINT (7.26789 6.15814) | 95 |
| 96 | POINT (5.885 6.05004) | 107 |
| 97 | POINT (3.54785 7.77382) | 146 |
| 98 | POINT (6.04603 3.09231) | 63 |
| 99 | POINT (2.45631 0.71266) | 60 |
100 rows × 2 columns
[8]:
fig, ax = plt.subplots(figsize=(4, 4))
_label = f"size-weighted clients\n\t$\\sum$={sum_clients}"
clients.plot(ax=ax, label=_label, alpha=0.3, markersize=ai * 4)
_label = f"capacity-weighted facilities\n\t$\\sum$={sum_capacity}"
facilities.plot(ax=ax, label=_label, alpha=0.3, markersize=cj)
plt.legend(loc="upper left", bbox_to_anchor=(1.05, 1));
2. The classic \(p\)-median problem¶
The \(p\)-median problem is a classic, introduced in Hakimi (1965), seeks to minimize the maximum weights for siting \(p\) facilities. As an integer linear program (\(ILP\)), the \(p\)-median problem is formulated as:
\(\begin{array} \displaystyle \textbf{Minimize} & \displaystyle \sum_{i \in I}\sum_{j \in J}{a_i d_{ij} X_{ij}} &&& (1) \\ \displaystyle \textbf{Subject to:} & \displaystyle \sum_{j \in J}{X_{ij} = 1} & \forall i \in I && (2) \\ & \displaystyle \sum_{j \in J}{Y_{j} = p} &&& (3) \\ & X_{ij} \leq Y_{j} & \forall i \in I & \forall j \in J & (4) \\ & X_{ij} \in \{0,1\} & \forall i \in I & \forall j \in J & (5) \\ & Y_{j} \in \{0,1\} & \forall j \in J && (6) \\ \end{array}\)
\(\begin{array} \displaystyle \textbf{Where:}\\ & & \displaystyle i & \small = & \textrm{index referencing client/demand locations} \\ & & j & \small = & \textrm{index referencing potential facility sites} \\ & & d_{ij} & \small = & \textrm{shortest distance between } i \textrm{ and } j \\ & & p & \small = & \textrm{number of facilities to be located} \\ & & a_i & \small = & \textrm{service load or population demand at } i \\ & & X_{ij} & \small = & \begin{cases} 1, \textrm{if demand } i \textrm{ is assigned to facility } j \\ 0, \textrm{otherwise} \end{cases} \\ & & Y_{j} & \small = & \begin{cases} 1, \textrm{if node } j \textrm{ has been selected for a facility} \\ 0, \textrm{otherwise} \\ \end{cases} \\ \end{array}\)
The formulation above is adapted from Church and Murray (2018)
First, we declare model positional and keyword arguments, which will be used in all \(p\)-median problem variants.
[9]:
model_args = clients, facilities, "geometry", "geometry", "weights", P_FAC
Instantiate and solve the classic \(p\)-median problem.
[10]:
pmedian_classic = PMedian.from_geodataframe(*model_args, name="classic-p-median")
pmedian_classic = pmedian_classic.solve(solver)
Record decision variable names used for mapping later.
[11]:
def fac_vars(pmp):
return [f.name.replace("_", "") for f in pmp.fac_vars]
[12]:
facilities["dv"] = fac_vars(pmedian_classic)
facilities = facilities[["geometry", "dv", "capacity"]].copy()
facilities
[12]:
| geometry | dv | capacity | |
|---|---|---|---|
| 0 | POINT (2.21993 8.70732) | y0 | 87 |
| 1 | POINT (2.06719 9.18611) | y1 | 190 |
| 2 | POINT (4.88411 6.11744) | y2 | 122 |
| 3 | POINT (7.65908 5.18418) | y3 | 187 |
| 4 | POINT (2.96801 1.87721) | y4 | 183 |
| ... | ... | ... | ... |
| 95 | POINT (7.26789 6.15814) | y95 | 95 |
| 96 | POINT (5.885 6.05004) | y96 | 107 |
| 97 | POINT (3.54785 7.77382) | y97 | 146 |
| 98 | POINT (6.04603 3.09231) | y98 | 63 |
| 99 | POINT (2.45631 0.71266) | y99 | 60 |
100 rows × 3 columns
Record solution diagnostics for model comparison.
[13]:
def pmp_diagnostic(pmp, add_to=None):
"""Helper for displaying model diagnostics."""
_diags = {
"model": pmp.name,
"variables": len(pmp.problem.variables()),
"constraints": len(pmp.problem.constraints),
"obj_val": round(pmp.problem.objective.value(), 3),
"selected": [f.name.replace("_", "") for f in pmp.fac_vars if f.varValue],
"cpu_time": pmp.problem.solutionCpuTime,
"wall_time": pmp.problem.solutionTime,
}
_diags = pandas.DataFrame([_diags.values()], columns=_diags.keys())
if isinstance(add_to, pandas.DataFrame):
_diags = pandas.concat([add_to, _diags], ignore_index=True)
return _diags
[14]:
diagnostic = pmp_diagnostic(pmedian_classic)
diagnostic
[14]:
| model | variables | constraints | obj_val | selected | cpu_time | wall_time | |
|---|---|---|---|---|---|---|---|
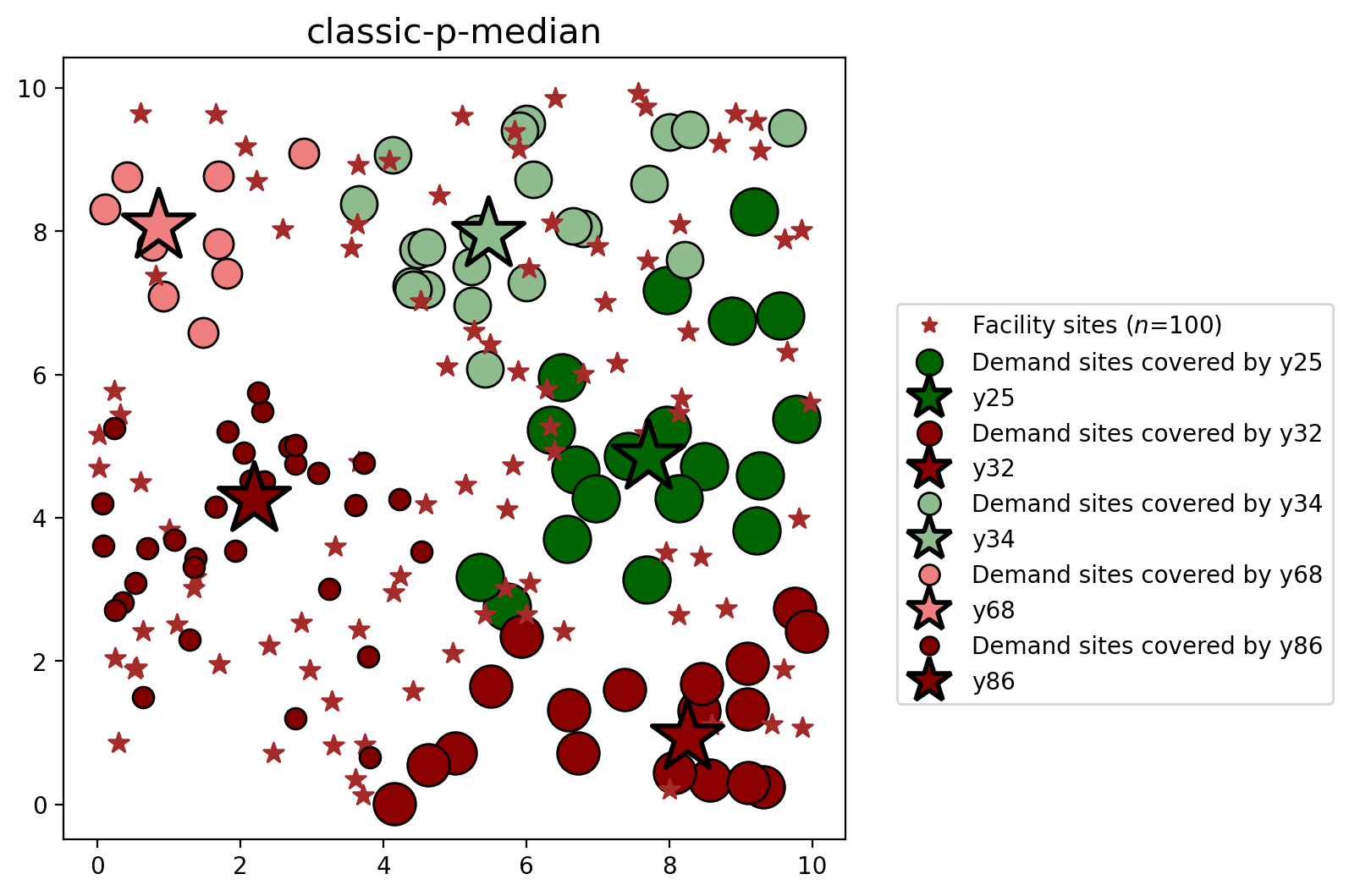
| 0 | classic-p-median | 10100 | 10101 | 668.88 | [y25, y34, y45, y68, y70, y88, y89, y91] | 0.272112 | 0.272112 |
The classic \(p\)-median problem does not take facility capacity into account, so we can (naively) assume that 100% of the capacity at each selected candidate facility is being utilized.
[15]:
def cli_sum(f2c):
return clients.loc[f2c, "weights"].sum()
def fac_cap(fdv):
return facilities.loc[(facilities["dv"] == fdv), "capacity"].squeeze()
def serv_perc_cap(f2c, fdv):
return round(cli_sum(f2c) / fac_cap(fdv), 2) * 100.0
def service_level(pmp):
if pmp.name.startswith("capacitated"):
zipped_vars = zip(pmp.fac2cli, facilities["dv"], strict=True)
serv_lev = [serv_perc_cap(f2c, fdv) for f2c, fdv in zipped_vars]
else:
serv_lev = [100.0 if bool(dv.varValue) else 0.0 for dv in pmp.fac_vars]
return serv_lev
[16]:
facilities["pmp_classic"] = service_level(pmedian_classic)
perc_cols = [c for c in facilities.columns if c.startswith("pmp")]
facilities.loc[(facilities[perc_cols] > 0).any(axis=1)]
[16]:
| geometry | dv | capacity | pmp_classic | |
|---|---|---|---|---|
| 25 | POINT (7.70854 4.84931) | y25 | 72 | 100.0 |
| 34 | POINT (5.46358 7.96143) | y34 | 80 | 100.0 |
| 45 | POINT (8.59707 1.11111) | y45 | 102 | 100.0 |
| 68 | POINT (0.8507 8.07777) | y68 | 73 | 100.0 |
| 70 | POINT (8.14642 8.10286) | y70 | 137 | 100.0 |
| 88 | POINT (3.65333 4.77941) | y88 | 126 | 100.0 |
| 89 | POINT (5.4209 2.6523) | y89 | 93 | 100.0 |
| 91 | POINT (1.3648 3.177) | y91 | 80 | 100.0 |
Plotting the results
[17]:
dv_colors_arr = list(mcolors.CSS4_COLORS.keys())
dv_colors = {f"y{i}": dv_colors_arr[i] for i in range(len(dv_colors_arr))}
[18]:
def plot_results(model, p, facs, clis=None):
"""Visualize optimal solution sets and context."""
fig, ax = plt.subplots(figsize=(6, 6))
markersize, markersize_factor = 4, 4
ax.set_title(model.name, fontsize=15)
# extract facility-client relationships for plotting
cli_points = {}
fac_sites = {}
for i, dv in enumerate(model.fac_vars):
if dv.varValue:
dv = facs.loc[i, "dv"]
fac_sites[dv] = i
geom = clis.iloc[model.fac2cli[i]]["geometry"]
cli_points[dv] = geom
# study area and legend entries initialization
legend_elements = []
# all candidate facilities
facs.plot(ax=ax, fc="brown", marker="*", markersize=80, zorder=8)
_label = f"Facility sites ($n$={len(model.fac_vars)})"
_mkws = {"marker": "*", "markerfacecolor": "brown", "markeredgecolor": "brown"}
legend_elements.append(mlines.Line2D([], [], ms=7, lw=0, label=_label, **_mkws))
# facility-(client) symbology and legend entries
zorder = 4
for fname, fac in fac_sites.items():
cset = dv_colors[fname]
# clients
geoms = cli_points[fname]
gdf = geopandas.GeoDataFrame(geoms)
gdf.plot(ax=ax, zorder=zorder, ec="k", fc=cset, markersize=100 * markersize)
_label = f"Demand sites covered by {fname}"
_mkws = {"markerfacecolor": cset, "markeredgecolor": "k", "ms": markersize + 7}
legend_elements.append(
mlines.Line2D([], [], marker="o", lw=0, label=_label, **_mkws)
)
# facilities
ec = "k"
lw = 2
facs.iloc[[fac]].plot(
ax=ax, marker="*", markersize=1000, zorder=9, fc=cset, ec=ec, lw=lw
)
_mkws = {"markerfacecolor": cset, "markeredgecolor": ec, "markeredgewidth": lw}
legend_elements.append(
mlines.Line2D([], [], marker="*", ms=20, lw=0, label=fname, **_mkws)
)
# increment zorder up and markersize down for stacked client symbology
zorder += 1
markersize -= markersize_factor / p
# legend
kws = {"loc": "upper left", "bbox_to_anchor": (1.05, 0.7)}
plt.legend(handles=legend_elements, **kws)
[19]:
plot_results(pmedian_classic, P_FAC, facilities, clis=clients)
3. The capacited \(p\)-median problem¶
The capacitated variant of the \(p\)-median problem is formulated identically to the classic \(p\)-median problem with the addition of contraint \((4)\) below. Here constraint \((4)\) stipulates that for a facility to be considered for selection it must have the avaialable capacity to support demand from allocated client locations. The capacitated \(p\)-median variant can be formulated as:
\(\begin{array} \displaystyle \textbf{Minimize} & \displaystyle \sum_{i \in I}\sum_{j \in J}{a_i d_{ij} X_{ij}} &&& (1) \\ \displaystyle \textbf{Subject to:} & \displaystyle \sum_{j \in J}{X_{ij} = 1} & \forall i \in I && (2) \\ & \displaystyle \sum_{j \in J}{Y_{j} = p} &&& (3) \\ & \displaystyle \sum_{i \in I}{a_i X_{ij} \leq {c_j Y_{j}}}& \forall j \in J && (4) \\ & X_{ij} \leq Y_{j} & \forall i \in I & \forall j \in J & (5) \\ & X_{ij} \in \{0,1\} & \forall i \in I & \forall j \in J & (6) \\ & Y_{j} \in \{0,1\} & \forall j \in J && (7) \\ \end{array}\)
\(\begin{array} \displaystyle \textbf{Where:}\\ & & \displaystyle i & \small = & \textrm{index referencing client/demand locations} \\ & & j & \small = & \textrm{index referencing potential facility sites} \\ & & d_{ij} & \small = & \textrm{shortest distance between } i \textrm{ and } j \\ & & p & \small = & \textrm{number of facilities to be located} \\ & & a_i & \small = & \textrm{service load or population demand at } i \\ & & c_j & \small = & \textrm{capacity of facility } j \\ & & X_{ij} & \small = & \begin{cases} 1, \textrm{if demand } i \textrm{ is assigned to facility } j \\ 0, \textrm{otherwise} \end{cases} \\ & & Y_{j} & \small = & \begin{cases} 1, \textrm{if node } j \textrm{ has been selected for a facility} \\ 0, \textrm{otherwise} \\ \end{cases} \\ \end{array}\)
The formulation above is adapted from Church and Murray (2009)
[20]:
pmedian_capacity = PMedian.from_geodataframe(
*model_args, name="p-median", facility_capacity_col="capacity"
)
pmedian_capacity = pmedian_capacity.solve(solver)
[21]:
diagnostic = pmp_diagnostic(pmedian_capacity, add_to=diagnostic)
diagnostic
[21]:
| model | variables | constraints | obj_val | selected | cpu_time | wall_time | |
|---|---|---|---|---|---|---|---|
| 0 | classic-p-median | 10100 | 10101 | 668.880 | [y25, y34, y45, y68, y70, y88, y89, y91] | 0.272112 | 0.272112 |
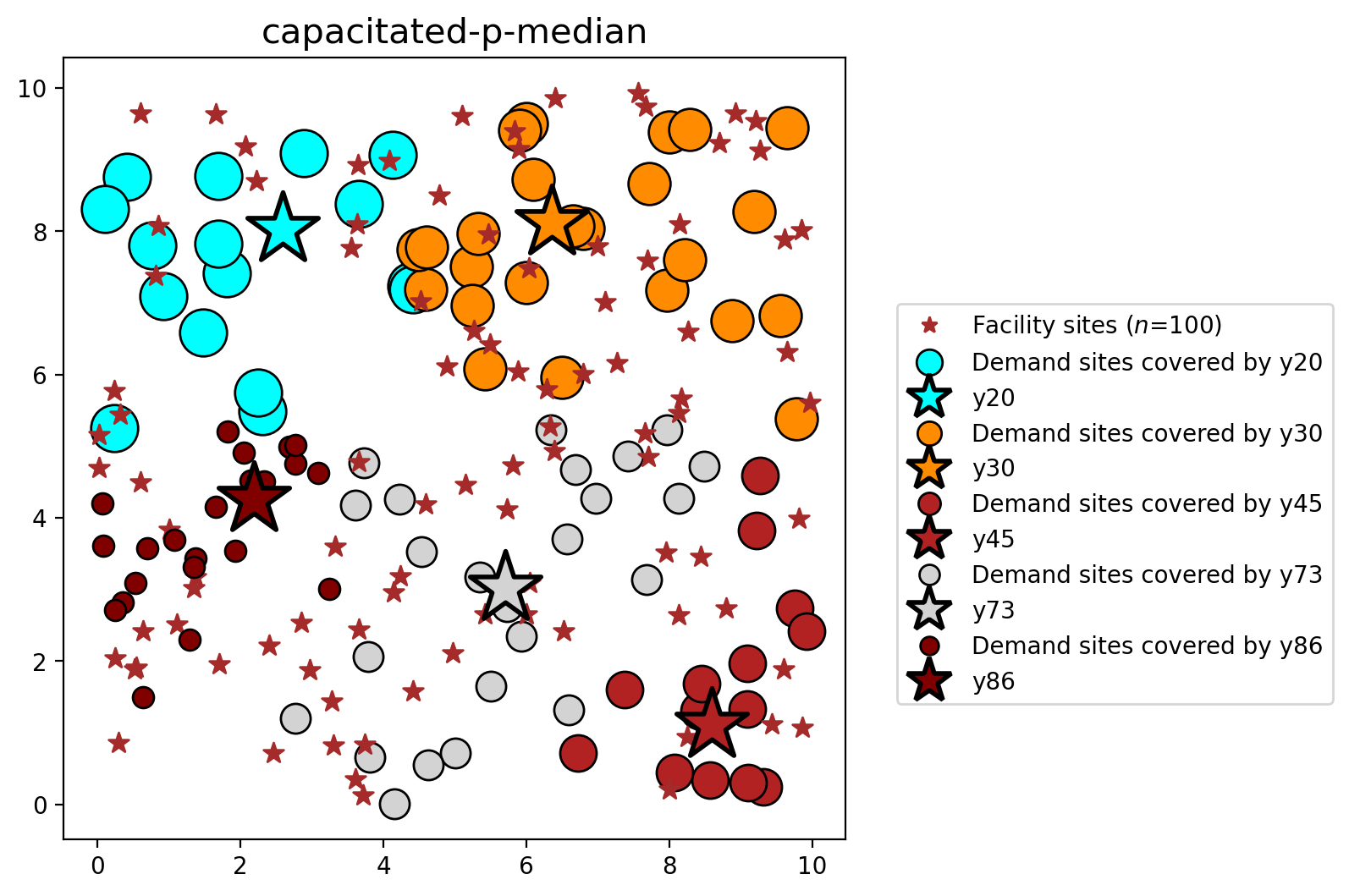
| 1 | capacitated-p-median | 10100 | 10201 | 675.964 | [y25, y45, y46, y68, y70, y88, y89, y91] | 0.409628 | 0.409629 |
[22]:
facilities["pmp_capacitated"] = service_level(pmedian_capacity)
perc_cols = [c for c in facilities.columns if c.startswith("pmp")]
facilities.loc[(facilities[perc_cols] > 0).any(axis=1)]
[22]:
| geometry | dv | capacity | pmp_classic | pmp_capacitated | |
|---|---|---|---|---|---|
| 25 | POINT (7.70854 4.84931) | y25 | 72 | 100.0 | 86.0 |
| 34 | POINT (5.46358 7.96143) | y34 | 80 | 100.0 | 0.0 |
| 45 | POINT (8.59707 1.11111) | y45 | 102 | 100.0 | 83.0 |
| 46 | POINT (4.78339 8.4998) | y46 | 130 | 0.0 | 80.0 |
| 68 | POINT (0.8507 8.07777) | y68 | 73 | 100.0 | 73.0 |
| 70 | POINT (8.14642 8.10286) | y70 | 137 | 100.0 | 50.0 |
| 88 | POINT (3.65333 4.77941) | y88 | 126 | 100.0 | 62.0 |
| 89 | POINT (5.4209 2.6523) | y89 | 93 | 100.0 | 63.0 |
| 91 | POINT (1.3648 3.177) | y91 | 80 | 100.0 | 86.0 |
[23]:
plot_results(pmedian_capacity, P_FAC, facilities, clis=clients)
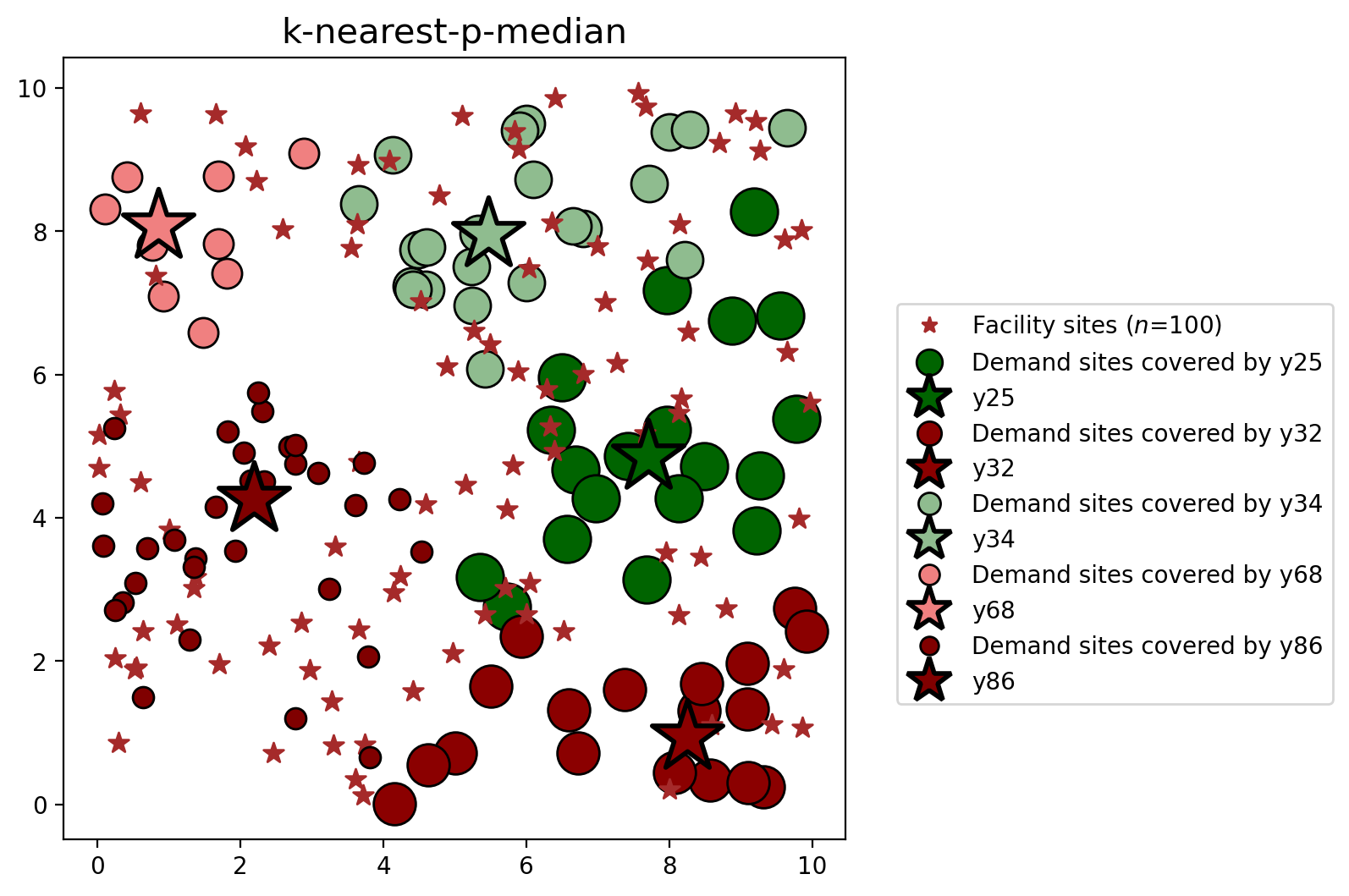
4. The \(k\)-nearest \(p\)-median problem¶
The \(k\)-nearest \(p\)-median problem, also referred to as the \(p\)-median model with near-far cost allocation, implements a variant of the classic \(p\)-median problem that only considers a limited selection of the nearest facilites, and was introduced in Church (2018). The focus here is the isolation a computationally efficient program for solving a \(p\)-median problem by dramatically reducing the number of variables and contraints in the mathematical formulation. As stated in the introduction above, a more detailed explanation of this model and its implementation can be found here. The \(k\)-nearest \(p\)-median variant can be formulated as:
\(\begin{array} \displaystyle \textbf{Minimize} & \displaystyle \sum_{i \in I}\sum_{k \in k_{i}}{a_i d_{ik} X_{ik}} + \sum_{i \in I}{g_i (d_{i{k_i}} + 1)} &&& (1) \\ \displaystyle \textbf{Subject to:} & \displaystyle \sum_{k \in k_{i}}{X_{ik} + g_i = 1} & \forall i \in I && (2) \\ & \displaystyle \sum_{j \in J}{Y_{j} = p} &&& (3) \\ & X_{ij} \leq Y_{j} & \forall i \in I & \forall j \in J & (4) \\ & X_{ij} \in \{0,1\} & \forall i \in I & \forall j \in J & (5) \\ & Y_{j} \in \{0,1\} & \forall j \in J && (6) \\ \end{array}\)
\(\begin{array} \displaystyle \textbf{Where:}\\ & & \displaystyle i & \small = & \textrm{index referencing client/demand locations} \\ & & j & \small = & \textrm{index referencing potential facility sites} \\ & & p & \small = & \textrm{number of facilities to be located} \\ & & a_i & \small = & \textrm{service load or population demand at } i \\ & & k_i & \small = & \textrm{the } k \textrm{ nearest facilities of client location } i \\ & & c_j & \small = & \textrm{capacity of facility } j \\ & & d_{ij} & \small = & \textrm{shortest distance between } i \textrm{ and } j \\ & & X_{ij} & \small = & \begin{cases} 1, \textrm{if demand } i \textrm{ is assigned to facility } j \\ 0, \textrm{otherwise} \end{cases} \\ & & Y_{j} & \small = & \begin{cases} 1, \textrm{if node } j \textrm{ has been selected for a facility} \\ 0, \textrm{otherwise} \\ \end{cases} \\ & & g_{i} & \small = & \begin{cases} 1, \textrm{if the client } i \textrm{ needs to be served by non-}k\textrm{-nearest facilities} \\ 0, \textrm{otherwise} \\ \end{cases} \\ \end{array}\)
The formulation above is adapted from Church (2018)
[24]:
pmedian_k_nearest = KNearestPMedian.from_geodataframe(
*model_args,
name="k-nearest-p-median",
)
pmedian_k_nearest = pmedian_k_nearest.solve(solver)
[25]:
diagnostic = pmp_diagnostic(pmedian_k_nearest, add_to=diagnostic)
diagnostic
[25]:
| model | variables | constraints | obj_val | selected | cpu_time | wall_time | |
|---|---|---|---|---|---|---|---|
| 0 | classic-p-median | 10100 | 10101 | 668.880 | [y25, y34, y45, y68, y70, y88, y89, y91] | 0.272112 | 0.272112 |
| 1 | capacitated-p-median | 10100 | 10201 | 675.964 | [y25, y45, y46, y68, y70, y88, y89, y91] | 0.409628 | 0.409629 |
| 2 | k-nearest-p-median | 1001 | 902 | 668.880 | [y25, y34, y45, y68, y70, y88, y89, y91] | 0.035709 | 0.035709 |
[26]:
facilities["pmp_k-nearest"] = service_level(pmedian_k_nearest)
perc_cols = [c for c in facilities.columns if c.startswith("pmp")]
facilities.loc[(facilities[perc_cols] > 0).any(axis=1)]
[26]:
| geometry | dv | capacity | pmp_classic | pmp_capacitated | pmp_k-nearest | |
|---|---|---|---|---|---|---|
| 25 | POINT (7.70854 4.84931) | y25 | 72 | 100.0 | 86.0 | 100.0 |
| 34 | POINT (5.46358 7.96143) | y34 | 80 | 100.0 | 0.0 | 100.0 |
| 45 | POINT (8.59707 1.11111) | y45 | 102 | 100.0 | 83.0 | 100.0 |
| 46 | POINT (4.78339 8.4998) | y46 | 130 | 0.0 | 80.0 | 0.0 |
| 68 | POINT (0.8507 8.07777) | y68 | 73 | 100.0 | 73.0 | 100.0 |
| 70 | POINT (8.14642 8.10286) | y70 | 137 | 100.0 | 50.0 | 100.0 |
| 88 | POINT (3.65333 4.77941) | y88 | 126 | 100.0 | 62.0 | 100.0 |
| 89 | POINT (5.4209 2.6523) | y89 | 93 | 100.0 | 63.0 | 100.0 |
| 91 | POINT (1.3648 3.177) | y91 | 80 | 100.0 | 86.0 | 100.0 |
[27]:
plot_results(pmedian_k_nearest, P_FAC, facilities, clis=clients)
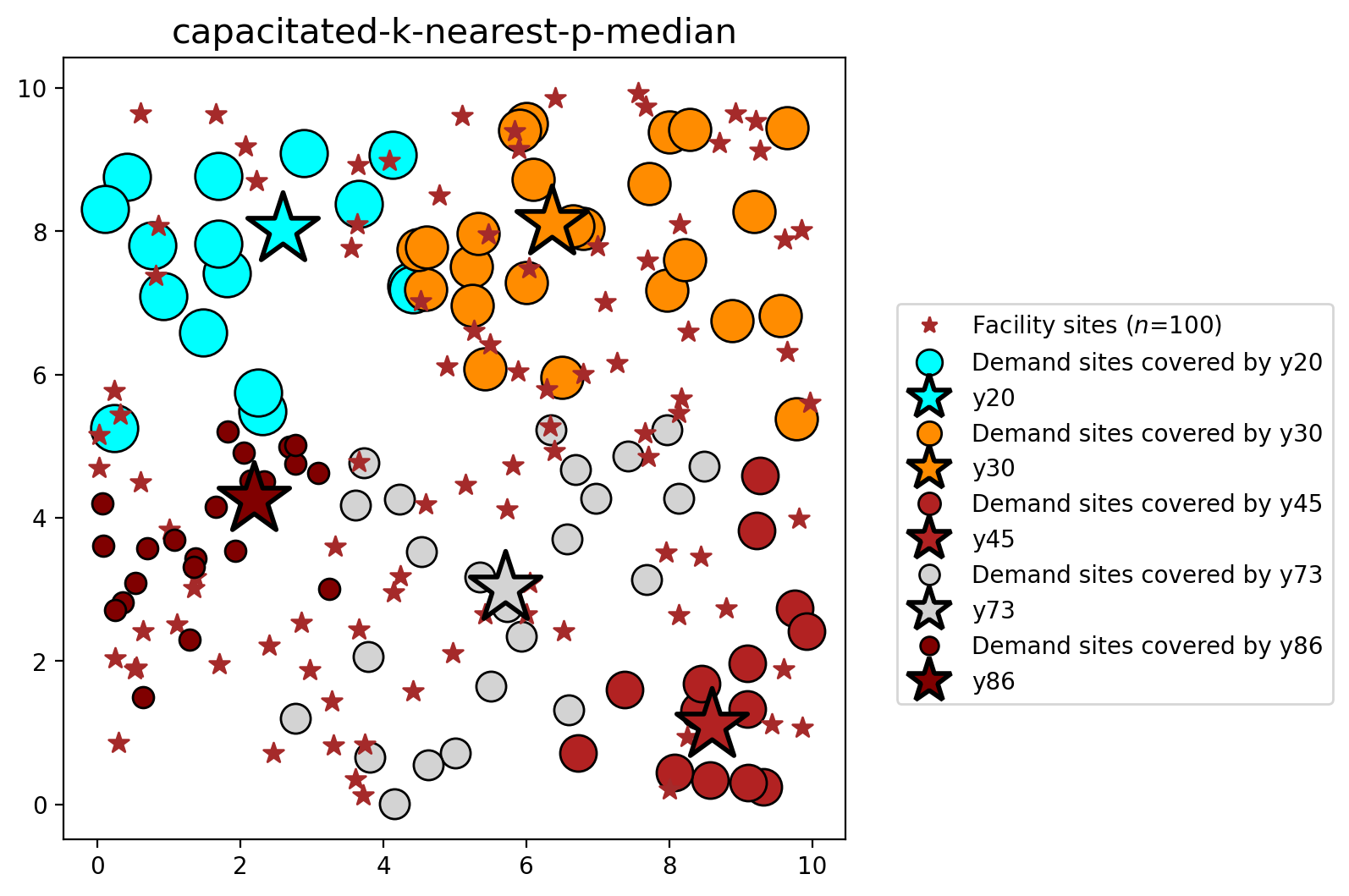
5. The capacited \(k\)-nearest \(p\)-median problem¶
With the addition of equation \((4)\) below, we can also introduce facility capacity as a constraint.
\(\begin{array} \displaystyle \textbf{Minimize} & \displaystyle \sum_{i \in I} \displaystyle \sum_{k \in k_{i}}{a_i d_{ik} X_{ik}} + \sum_{i \in I}{g_i (d_{i{k_i}} + 1)} &&& (1) \\ \displaystyle \textbf{Subject to:} & \displaystyle \sum_{k \in k_{i}}{X_{ik} + g_i = 1} & \forall i \in I && (2) \\ & \displaystyle \sum_{j \in J}{Y_{j} = p} &&& (3) \\ & \displaystyle \sum_{i \in I}{a_i X_{ij} \leq {c_j Y_{j}}}& \forall j \in J && (4) \\ & X_{ij} \leq Y_{j} & \forall i \in I & \forall j \in J & (5) \\ & X_{ij} \in \{0,1\} & \forall i \in I & \forall j \in J & (6) \\ & Y_{j} \in \{0,1\} & \forall j \in J && (7) \\ \end{array}\)
\(\begin{array} \displaystyle \textbf{Where:}\\ & & \displaystyle i & \small = & \textrm{index referencing client/demand locations} \\ & & j & \small = & \textrm{index referencing potential facility sites} \\ & & p & \small = & \textrm{number of facilities to be located} \\ & & a_i & \small = & \textrm{service load or population demand at } i \\ & & k_i & \small = & \textrm{the } k^{th} \textrm{nearest facilities of client location } i \\ & & c_j & \small = & \textrm{capacity of facility } j \\ & & d_{ij} & \small = & \textrm{shortest distance between } i \textrm{ and } j \\ & & X_{ij} & \small = & \begin{cases} 1, \textrm{if demand } i \textrm{ is assigned to facility } j \\ 0, \textrm{otherwise} \end{cases} \\ & & Y_{j} & \small = & \begin{cases} 1, \textrm{if node } j \textrm{ has been selected for a facility} \\ 0, \textrm{otherwise} \\ \end{cases} \\ & & g_{i} & \small = & \begin{cases} 1, \textrm{if the client } i \textrm{ needs to be served by non-k-nearest facilities} \\ 0, \textrm{otherwise} \\ \end{cases} \\ \end{array}\)
The formulation above is adapted from Church and Murray (2018)
[28]:
pmedian_k_nearest_capacity = KNearestPMedian.from_geodataframe(
*model_args,
name="k-nearest-p-median",
facility_capacity_col="capacity",
)
pmedian_k_nearest_capacity = pmedian_k_nearest_capacity.solve(solver)
[29]:
diagnostic = pmp_diagnostic(pmedian_k_nearest_capacity, add_to=diagnostic)
diagnostic
[29]:
| model | variables | constraints | obj_val | selected | cpu_time | wall_time | |
|---|---|---|---|---|---|---|---|
| 0 | classic-p-median | 10100 | 10101 | 668.880 | [y25, y34, y45, y68, y70, y88, y89, y91] | 0.272112 | 0.272112 |
| 1 | capacitated-p-median | 10100 | 10201 | 675.964 | [y25, y45, y46, y68, y70, y88, y89, y91] | 0.409628 | 0.409629 |
| 2 | k-nearest-p-median | 1001 | 902 | 668.880 | [y25, y34, y45, y68, y70, y88, y89, y91] | 0.035709 | 0.035709 |
| 3 | capacitated-k-nearest-p-median | 1067 | 1835 | 675.964 | [y25, y45, y46, y68, y70, y88, y89, y91] | 0.379421 | 0.379421 |
[30]:
facilities["pmp_k-nearest_capacitated"] = service_level(pmedian_k_nearest_capacity)
perc_cols = [c for c in facilities.columns if c.startswith("pmp")]
facilities.loc[(facilities[perc_cols] > 0).any(axis=1)]
[30]:
| geometry | dv | capacity | pmp_classic | pmp_capacitated | pmp_k-nearest | pmp_k-nearest_capacitated | |
|---|---|---|---|---|---|---|---|
| 25 | POINT (7.70854 4.84931) | y25 | 72 | 100.0 | 86.0 | 100.0 | 86.0 |
| 34 | POINT (5.46358 7.96143) | y34 | 80 | 100.0 | 0.0 | 100.0 | 0.0 |
| 45 | POINT (8.59707 1.11111) | y45 | 102 | 100.0 | 83.0 | 100.0 | 83.0 |
| 46 | POINT (4.78339 8.4998) | y46 | 130 | 0.0 | 80.0 | 0.0 | 80.0 |
| 68 | POINT (0.8507 8.07777) | y68 | 73 | 100.0 | 73.0 | 100.0 | 73.0 |
| 70 | POINT (8.14642 8.10286) | y70 | 137 | 100.0 | 50.0 | 100.0 | 50.0 |
| 88 | POINT (3.65333 4.77941) | y88 | 126 | 100.0 | 62.0 | 100.0 | 62.0 |
| 89 | POINT (5.4209 2.6523) | y89 | 93 | 100.0 | 63.0 | 100.0 | 63.0 |
| 91 | POINT (1.3648 3.177) | y91 | 80 | 100.0 | 86.0 | 100.0 | 86.0 |
[31]:
plot_results(pmedian_k_nearest_capacity, P_FAC, facilities, clis=clients)
6. Comparing variant results¶
Solution diagnostics & optimal facility selection¶
Below we can see that:
The classic \(p\)-median and \(k\)-nearest variant result in equivalent objective values and facility selections. However, the \(k\)-nearest variant achieves optimality from a formulation with approximately 15% the number of variables and constraints of the classic formulation, resulting in a dramatically reduced solution time.
Similar results are observed with the capacitated variants: equivalent objective values and facility selections, plus solve times approximately 20% faster.
Caveat: The solve times for the \(k\)-nearest variants do not include preparation of the minimal formulation, only actual solve time reported via
pulp.
[32]:
diagnostic
[32]:
| model | variables | constraints | obj_val | selected | cpu_time | wall_time | |
|---|---|---|---|---|---|---|---|
| 0 | classic-p-median | 10100 | 10101 | 668.880 | [y25, y34, y45, y68, y70, y88, y89, y91] | 0.272112 | 0.272112 |
| 1 | capacitated-p-median | 10100 | 10201 | 675.964 | [y25, y45, y46, y68, y70, y88, y89, y91] | 0.409628 | 0.409629 |
| 2 | k-nearest-p-median | 1001 | 902 | 668.880 | [y25, y34, y45, y68, y70, y88, y89, y91] | 0.035709 | 0.035709 |
| 3 | capacitated-k-nearest-p-median | 1067 | 1835 | 675.964 | [y25, y45, y46, y68, y70, y88, y89, y91] | 0.379421 | 0.379421 |
[33]:
facilities.loc[(facilities[perc_cols] > 0).any(axis=1)]
[33]:
| geometry | dv | capacity | pmp_classic | pmp_capacitated | pmp_k-nearest | pmp_k-nearest_capacitated | |
|---|---|---|---|---|---|---|---|
| 25 | POINT (7.70854 4.84931) | y25 | 72 | 100.0 | 86.0 | 100.0 | 86.0 |
| 34 | POINT (5.46358 7.96143) | y34 | 80 | 100.0 | 0.0 | 100.0 | 0.0 |
| 45 | POINT (8.59707 1.11111) | y45 | 102 | 100.0 | 83.0 | 100.0 | 83.0 |
| 46 | POINT (4.78339 8.4998) | y46 | 130 | 0.0 | 80.0 | 0.0 | 80.0 |
| 68 | POINT (0.8507 8.07777) | y68 | 73 | 100.0 | 73.0 | 100.0 | 73.0 |
| 70 | POINT (8.14642 8.10286) | y70 | 137 | 100.0 | 50.0 | 100.0 | 50.0 |
| 88 | POINT (3.65333 4.77941) | y88 | 126 | 100.0 | 62.0 | 100.0 | 62.0 |
| 89 | POINT (5.4209 2.6523) | y89 | 93 | 100.0 | 63.0 | 100.0 | 63.0 |
| 91 | POINT (1.3648 3.177) | y91 | 80 | 100.0 | 86.0 | 100.0 | 86.0 |